Andrew Donson, Senior Research Associate
The Morgan Adams Foundation Pediatric Brain Tumor Research Program
at the University of Colorado Anschutz Medical Campus & Children’s Hospital Colorado
Andy Donson has been an integral member of the research team since the lab first opened its doors more than 25 years ago. “I was a biochemistry major in England, and I started working with Nick Foreman in 1993 at the University of Bristol. Nick was hired to start a neuro-oncology program at Children’s Hospital Colorado in 1995, and I followed him here.”
Read below for Andy’s explanation of how advancing technology has enhanced the genetic information that can be obtained from the study of pediatric cancer tumor samples.
The evolution of genetic sequencing: from “chips” to RNA sequencing and single-cell analysis
How it all started
Originally, it was just me in the lab, and Nick was treating patients. We were the only people in the region treating neuro-oncology patients at the time. Then, the Morgan Adams factor kicked in. They began working with us in 1999 and when we started gene research in 2004, MAF funded our first “gene chipping” project. We had already started banking samples from tumors, so we had a great foundation to start from. We started getting more data, becoming more successful, getting more papers published, and establishing a presence in the brain tumor research community.
The early years of genetic research
The Morgan Adams Foundation has advanced our understanding of childhood brain tumors through the use of “chipping”, where we simultaneously measured the expression of every single gene in a child’s cancer. In this process, we took a crumb-sized fragment of a patient’s tumor specimen removed during surgery and extracted all of the RNA contained in that fragment. A small piece of tumor contains around one million cells, so the data that we obtained by chipping a sample was the average of those million cells.
Ten years ago, we used chipping to ask the question “what genes distinguish tumors of children who are cured from the tumors that grow back?” The answer was that the tumors that were cured had higher immune gene expression.
Following up on this surprising result, we discovered that a tumor is not simply comprised of tumor cells but also immune cells. We also found that the ratio of immune cells to tumor cells was higher in the patients who didn’t relapse compared to those whose tumors regrew. These two discoveries alerted us to the existence and important role of different cell types within every tumor. It also presented us with a new question of how to “chip” and learn more about individual cells in a tumor, which we were unable to answer at the time.
Evolving technology leads to new discoveries
Ten years later, “chipping” has been replaced by “RNA sequencing,” a highly versatile 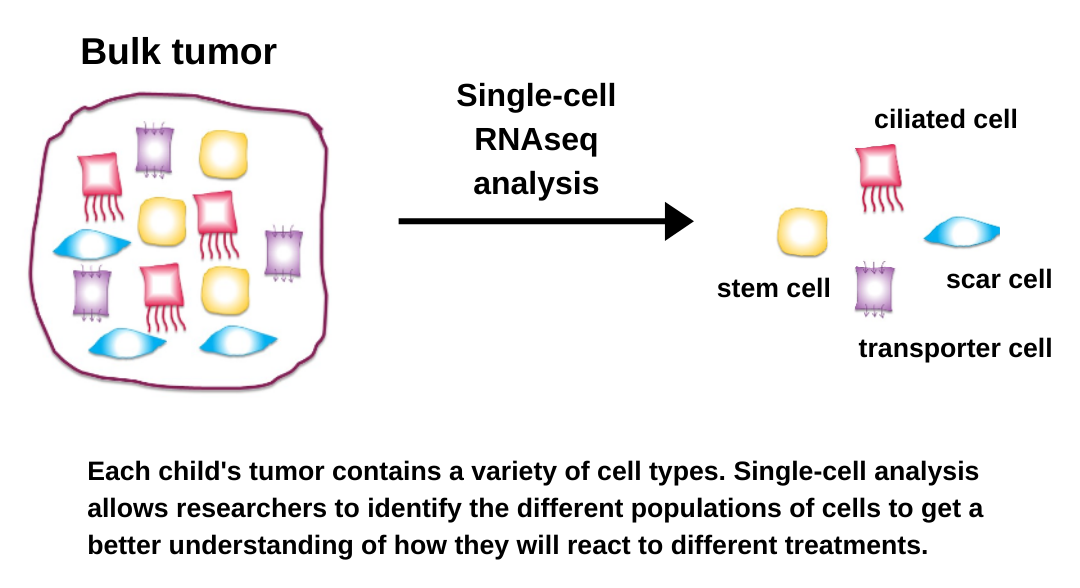 technology that now allows us to sequence the entire human genome in 2 days (compared to 5 years using the earlier technology!). Researchers combined this breakthrough technology with microscopic fluidics engineering and created a way to “chip” single cells – called single-cell RNA sequencing. The University of Colorado Anschutz Medical Campus quickly invested in this technology, allowing us to finally perform the experiments that we had dreamed up 10 years earlier.
technology that now allows us to sequence the entire human genome in 2 days (compared to 5 years using the earlier technology!). Researchers combined this breakthrough technology with microscopic fluidics engineering and created a way to “chip” single cells – called single-cell RNA sequencing. The University of Colorado Anschutz Medical Campus quickly invested in this technology, allowing us to finally perform the experiments that we had dreamed up 10 years earlier.
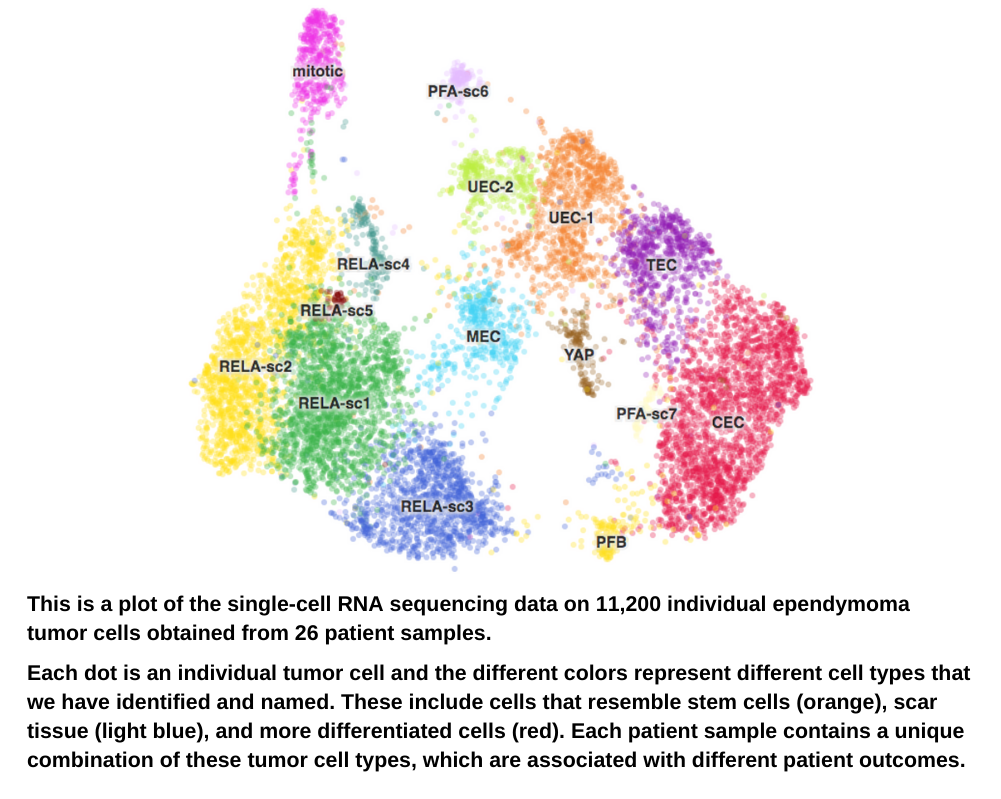
Over the past 2 years, we have used single-cell RNA sequencing to provide us with profound insights into childhood brain tumors that impact our understanding of the biology that drives these tumors. As a result, we have discovered promising new targets for therapeutics.
For all of the main childhood cancer types, we have identified and characterized multiple different mutated cancer cell types in addition to those immune cells that we saw in earlier studies. This includes cancer cell types with “stem-cell” properties, and we can now better focus our future efforts to target these more malignant subpopulations of tumor cells.
The goal of this work, as with everything we do, is to create more effective treatment options for kids and teens with cancer.